- Record: found
- Abstract: found
- Article: found
High-sensitivity profiling of SARS-CoV-2 noncoding region–host protein interactome reveals the potential regulatory role of negative-sense viral RNA
Read this article at
ABSTRACT
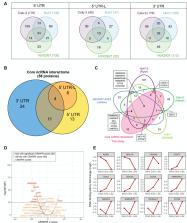
A deep understanding of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–host interactions is crucial to developing effective therapeutics and addressing the threat of emerging coronaviruses. The role of noncoding regions of viral RNA (ncrRNAs) has yet to be systematically scrutinized. We developed a method using MS2 affinity purification coupled with liquid chromatography-mass spectrometry and designed a diverse set of bait ncrRNAs to systematically map the interactome of SARS-CoV-2 ncrRNA in Calu-3, Huh7, and HEK293T cells. Integration of the results defined the core ncrRNA–host protein interactomes among cell lines. The 5′ UTR interactome is enriched with proteins in the small nuclear ribonucleoproteins family and is a target for the regulation of viral replication and transcription. The 3′ UTR interactome is enriched with proteins involved in the stress granules and heterogeneous nuclear ribonucleoproteins family. Intriguingly, compared with the positive-sense ncrRNAs, the negative-sense ncrRNAs, especially the negative-sense of 3′ UTR, interacted with a large array of host proteins across all cell lines. These proteins are involved in the regulation of the viral production process, host cell apoptosis, and immune response. Taken together, our study depicts the comprehensive landscape of the SARS-CoV-2 ncrRNA–host protein interactome and unveils the potential regulatory role of the negative-sense ncrRNAs, providing a new perspective on virus–host interactions and the design of future therapeutics. Given the highly conserved nature of UTRs in positive-strand viruses, the regulatory role of negative-sense ncrRNAs should not be exclusive to SARS-CoV-2.
IMPORTANCE
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes COVID-19, a pandemic affecting millions of lives. During replication and transcription, noncoding regions of the viral RNA (ncrRNAs) may play an important role in the virus–host interactions. Understanding which and how these ncrRNAs interact with host proteins is crucial for understanding the mechanism of SARS-CoV-2 pathogenesis. We developed the MS2 affinity purification coupled with liquid chromatography-mass spectrometry method and designed a diverse set of ncrRNAs to identify the SARS-CoV-2 ncrRNA interactome comprehensively in different cell lines and found that the 5′ UTR binds to proteins involved in U1 small nuclear ribonucleoprotein, while the 3′ UTR interacts with proteins involved in stress granules and the heterogeneous nuclear ribonucleoprotein family. Interestingly, negative-sense ncrRNAs showed interactions with a large number of diverse host proteins, indicating a crucial role in infection. The results demonstrate that ncrRNAs could serve diverse regulatory functions.
Related collections
Most cited references97
- Record: found
- Abstract: found
- Article: not found
Clinical Characteristics of Coronavirus Disease 2019 in China
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.
- Record: found
- Abstract: found
- Article: not found