- Record: found
- Abstract: found
- Article: found
Distinguishing retinal angiomatous proliferation from polypoidal choroidal vasculopathy with a deep neural network based on optical coherence tomography
Read this article at
Abstract
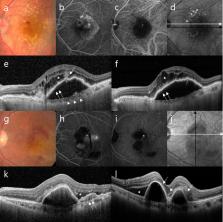
This cross-sectional study aimed to build a deep learning model for detecting neovascular age-related macular degeneration (AMD) and to distinguish retinal angiomatous proliferation (RAP) from polypoidal choroidal vasculopathy (PCV) using a convolutional neural network (CNN). Patients from a single tertiary center were enrolled from January 2014 to January 2020. Spectral-domain optical coherence tomography (SD-OCT) images of patients with RAP or PCV and a control group were analyzed with a deep CNN. Sensitivity, specificity, accuracy, and area under the receiver operating characteristic curve (AUROC) were used to evaluate the model’s ability to distinguish RAP from PCV. The performances of the new model, the VGG-16, Resnet-50, Inception, and eight ophthalmologists were compared. A total of 3951 SD-OCT images from 314 participants (229 AMD, 85 normal controls) were analyzed. In distinguishing the PCV and RAP cases, the proposed model showed an accuracy, sensitivity, and specificity of 89.1%, 89.4%, and 88.8%, respectively, with an AUROC of 95.3% (95% CI 0.727–0.852). The proposed model showed better diagnostic performance than VGG-16, Resnet-50, and Inception-V3 and comparable performance with the eight ophthalmologists. The novel model performed well when distinguishing between PCV and RAP. Thus, automated deep learning systems may support ophthalmologists in distinguishing RAP from PCV.
Related collections
Most cited references36
- Record: found
- Abstract: not found
- Article: not found
ImageNet classification with deep convolutional neural networks
- Record: found
- Abstract: found
- Article: found
A survey on Image Data Augmentation for Deep Learning

- Record: found
- Abstract: found
- Article: found