- Record: found
- Abstract: found
- Article: found
FangNet: Mining herb hidden knowledge from TCM clinical effective formulas using structure network algorithm

Read this article at
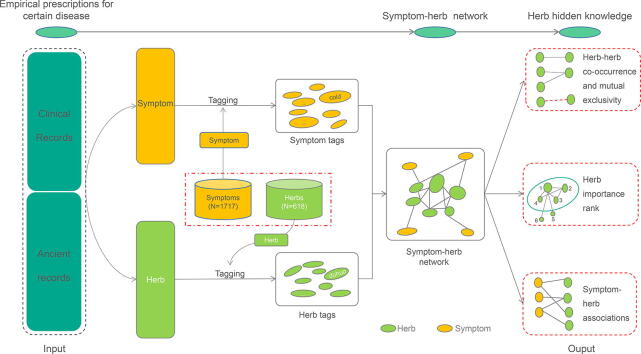
Graphical abstract
Abstract
The use of herbs to treat various human diseases has been recorded for thousands of years. In Asia's current medical system, numerous herbal formulas have been repeatedly verified to confirm their effectiveness in different periods, which is a great resource for drug innovation and discovery. Through the mining of these clinical effective formulas by network pharmacology and bioinformatics analysis, important biologically active ingredients derived from these natural products might be discovered. As modern medicine requires a combination of multiple drugs for the treatment of complex diseases, previously clinical formulas are also combinations of various herbs according to the main causes and accompanying symptoms. However, the herbs that play a major role in the treatment of diseases are always unclear. Therefore, how to rank each herb's relative importance and determine the core herbs, is the first step to assisting herb selection for active ingredients discovery. To solve this problem, we built the platform FangNet, which ranks all herbs on their relative topological importance using the PageRank algorithm, based on the constructed symptom-herb network from a collection of clinical empirical prescriptions. Three types of herb hidden knowledge, including herb importance rank, herb-herb co-occurrence, and associations to symptoms, were provided in an interactive visualization. Moreover, FangNet has designed role-based permission for teams to store, analyze, and jointly interpret their clinical formulas, in an easy and secure collaboration environment, aiming at creating a central hub for massive symptom-herb connections. FangNet can be accessed at http://fangnet.org or http://fangnet.herb.ac.cn.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Deep learning in neural networks: An overview
- Record: found
- Abstract: found
- Article: not found
The re-emergence of natural products for drug discovery in the genomics era.
- Record: found
- Abstract: found
- Article: not found