- Record: found
- Abstract: found
- Article: found
Ubiquitinome Profiling Reveals in Vivo UBE2D3 Targets and Implicates UBE2D3 in Protein Quality Control

Read this article at
Abstract
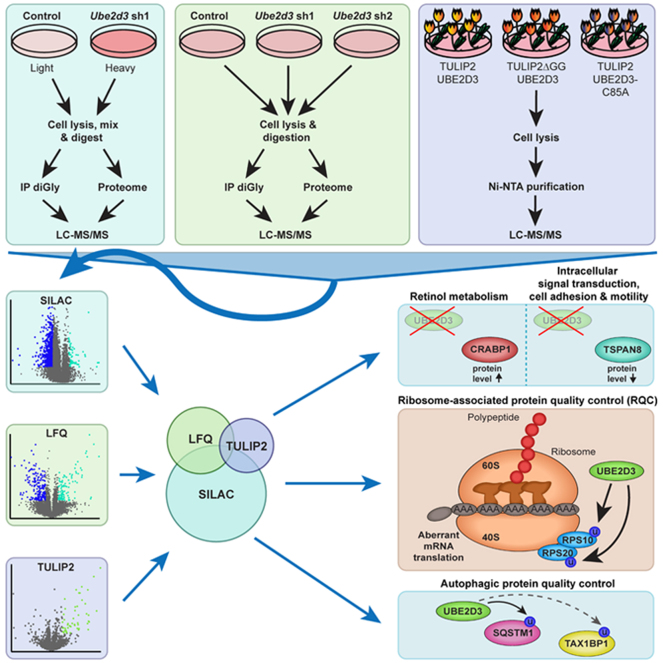
Ubiquitination has crucial roles in many cellular processes, and dysregulation of ubiquitin machinery enzymes can result in various forms of pathogenesis. Cells only have a limited set of ubiquitin-conjugating (E2) enzymes to support the ubiquitination of many cellular targets. As individual E2 enzymes have many different substrates and interactions between E2 enzymes and their substrates can be transient, it is challenging to define all in vivo substrates of an individual E2 and the cellular processes it affects. Particularly challenging in this respect is UBE2D3, an E2 enzyme with promiscuous activity in vitro but less defined roles in vivo. Here, we set out to identify in vivo targets of UBE2D3 by using stable isotope labeling by amino acids in cell culture–based and label-free quantitative ubiquitin diGly proteomics to study global proteome and ubiquitinome changes associated with UBE2D3 depletion. UBE2D3 depletion changed the global proteome, with the levels of proteins from metabolic pathways, in particular retinol metabolism, being the most affected. However, the impact of UBE2D3 depletion on the ubiquitinome was much more prominent. Interestingly, molecular pathways related to mRNA translation were the most affected. Indeed, we find that ubiquitination of the ribosomal proteins RPS10 and RPS20, critical for ribosome-associated protein quality control, is dependent on UBE2D3. We show by Targets of Ubiquitin Ligases Identified by Proteomics 2 methodology that RPS10 and RPS20 are direct targets of UBE2D3 and demonstrate that the catalytic activity of UBE2D3 is required to ubiquitinate RPS10 in vivo. In addition, our data suggest that UBE2D3 acts at multiple levels in autophagic protein quality control. Collectively, our findings show that depletion of an E2 enzyme in combination with quantitative diGly-based ubiquitinome profiling is a powerful tool to identify new in vivo E2 substrates, as we have done here for UBE2D3. Our work provides an important resource for further studies on the in vivo functions of UBE2D3.
Graphical Abstract
Highlights
In Brief
To identify in vivo substrates of the E2 ubiquitin-conjugating enzyme UBE2D3, we combined SILAC-based and label-free diGly-proteomics with UBE2D3 depletion. We show that UBE2D3 impacts CRABP1 and TSPAN8 levels and ubiquitinates key proteins in protein quality control. TULIP2 methodology identifies RPS10, RPS20, and SQSTM1 among direct targets of UBE2D3, and we show that UBE2D3's catalytic activity is required for RPS10 ubiquitination in vivo. Our approach identifies many potential UBE2D3 substrates, thereby providing a resource for future studies on UBE2D3 functions.
Related collections
Most cited references92

- Record: found
- Abstract: found
- Article: found
STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets

- Record: found
- Abstract: found
- Article: found
The PRIDE database and related tools and resources in 2019: improving support for quantification data
- Record: found
- Abstract: found
- Article: not found