- Record: found
- Abstract: found
- Article: found
Draft Genome Sequence of Streptomyces mexicanus Strain Q0842, Isolated from Human Skin
Read this article at
Abstract
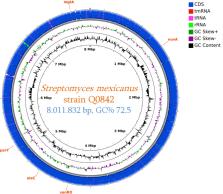
In 2003, Streptomyces mexicanus was reported as a novel xylanolytic bacterial species isolated from soil; a partial genome sequence was determined. In 2019, a strain from the same species was isolated from a hand skin swab sample from a healthy French woman. Genome sequencing revealed an 8,011,832-bp sequence with a GC content of 72.5%.
ABSTRACT
In 2003, Streptomyces mexicanus was reported as a novel xylanolytic bacterial species isolated from soil; a partial genome sequence was determined. In 2019, a strain from the same species was isolated from a hand skin swab sample from a healthy French woman. Genome sequencing revealed an 8,011,832-bp sequence with a GC content of 72.5%.
Related collections
Most cited references11

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.
- Record: found
- Abstract: found
- Article: not found