- Record: found
- Abstract: found
- Article: found
Variation in faecal microbiota in a group of horses managed at pasture over a 12-month period
Read this article at
Abstract
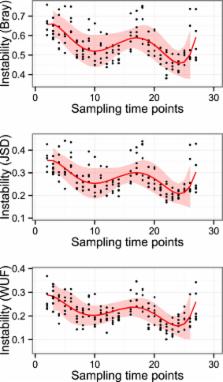
Colic (abdominal pain) is a common cause of mortality in horses. Change in management of horses is associated with increased colic risk and seasonal patterns of increased risk have been identified. Shifts in gut microbiota composition in response to management change have been proposed as one potential underlying mechanism for colic. However, the intestinal microbiota in normal horses and how this varies over different seasons has not previously been investigated. In this study the faecal microbiota composition was studied over 12 months in a population of horses managed at pasture with minimal changes in management. We hypothesised that gut microbiota would be stable in this population over time. Faecal samples were collected every 14 days from 7 horses for 52 weeks and the faecal microbiota was characterised by next-generation sequencing of 16S rRNA genes. The faecal microbiota was dominated by members of the phylum Firmicutes and Bacteroidetes throughout. Season, supplementary forage and ambient weather conditions were significantly associated with change in the faecal microbiota composition. These results provide important baseline information demonstrating physiologic variation in the faecal microbiota of normal horses over a 12-month period without development of colic.
Related collections
Most cited references32
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: not found
- Article: not found
Multiple Comparisons among Means
- Record: found
- Abstract: found
- Article: not found