- Record: found
- Abstract: found
- Article: not found
A small cell lung cancer genome reports complex tobacco exposure signatures
Read this article at
SUMMARY
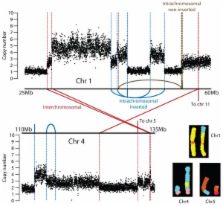
Cancer is driven by mutation. Worldwide, tobacco smoking is the major lifestyle exposure that causes cancer, exerting carcinogenicity through >60 chemicals that bind and mutate DNA. Using massively parallel sequencing technology, we sequenced a small cell lung cancer cell line, NCI-H209, to explore the mutational burden associated with tobacco smoking. 22,910 somatic substitutions were identified, including 132 in coding exons. Multiple mutation signatures testify to the cocktail of carcinogens in tobacco smoke and their proclivities for particular bases and surrounding sequence context. Effects of transcription-coupled repair and a second, more general expression-linked repair pathway were evident. We identified a tandem duplication that duplicates exons 3-8 of CHD7 in-frame, and another two lines carrying PVT1-CHD7 fusion genes, suggesting that CHD7 may be recurrently rearranged in this disease. These findings illustrate the potential for next-generation sequencing to provide unprecedented insights into mutational processes, cellular repair pathways and gene networks associated with cancer.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
DNA methylation profiling of human chromosomes 6, 20 and 22
- Record: found
- Abstract: found
- Article: not found
