- Record: found
- Abstract: found
- Article: not found
Retrotransposition-Competent Human LINE-1 Induces Apoptosis in Cancer Cells With Intact p53
Read this article at
Abstract
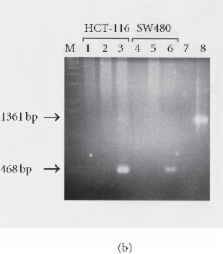
Retrotransposition of human LINE-1 (L1) element, a major representative non-LTR retrotransposon in the human genome, is known to be a source of insertional mutagenesis. However, nothing is known about effects of L1 retrotransposition on cell growth and differentiation. To investigate the potential for such biological effects and the impact that human L1 retrotransposition has upon cancer cell growth, we examined a panel of human L1 transformed cell lines following a complete retrotransposition process. The results demonstrated that transposition of L1 leads to the activation of the p53-mediated apoptotic pathway in human cancer cells that possess a wild-type p53. In addition, we found that inactivation of p53 in cells, where L1 was undergoing retrotransposition, inhibited the induction of apoptosis. This suggests an association between active retrotransposition and a competent p53 response in which induction of apoptosis is a major outcome. These data are consistent with a model in which human retrotransposition is sensed by the cell as a “genetic damaging event” and that massive retrotransposition triggers signaling pathways resulting in apoptosis.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Human LINE retrotransposons generate processed pseudogenes.
- Record: found
- Abstract: found
- Article: not found
Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man.
- Record: found
- Abstract: found
- Article: not found
