- Record: found
- Abstract: found
- Article: found
The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer
Read this article at
Abstract
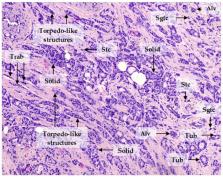
Intratumor morphological heterogeneity reflects patterns of invasive growth and is an indicator of the metastatic potential of breast cancer. In this study, we used this heterogeneity to identify molecules associated with breast cancer invasion and metastasis. The gene expression microarray data were used to identify genes differentially expressed between solid, trabecular, and other morphological arrangements of tumor cells. Immunohistochemistry was applied to evaluate the association of the selected proteins with metastasis. RNA-sequencing was performed to analyze the molecular makeup of metastatic tumor cells. High frequency of metastases and decreased metastasis-free survival were detected in patients either with positive expression of KIF14 or Mieap or negative expression of EZR at the tips of the torpedo-like structures in breast cancers. KIF14- and Mieap-positive and EZR-negative cells were mainly detected in the torpedo-like structures of the same breast tumors; however, their transcriptomic features differed. KIF14-positive cells showed a significant upregulation of genes involved in ether lipid metabolism. Mieap-positive cells were enriched in genes involved in mitophagy. EZR-negative cells displayed upregulated genes associated with phagocytosis and the chemokine-mediated signaling pathway. In conclusion, the positive expression of KIF14 and Mieap and negative expression of EZR at the tips of the torpedo-like structures are associated with breast cancer metastasis.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Analysis of the kinesin superfamily: insights into structure and function.
- Record: found
- Abstract: found
- Article: not found
Lung metastasis genes couple breast tumor size and metastatic spread.
- Record: found
- Abstract: found
- Article: not found