- Record: found
- Abstract: found
- Article: found
Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering
Read this article at
Abstract
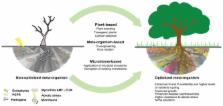
The goal of microbiome engineering is to manipulate the microbiome toward a certain type of community that will optimize plant functions of interest. For instance, in crop production the goal is to reduce disease susceptibility, increase nutrient availability increase abiotic stress tolerance and increase crop yields. Various approaches can be devised to engineer the plant–microbiome, but one particularly promising approach is to take advantage of naturally evolved plant–microbiome communication channels. This is, however, very challenging as the understanding of the plant–microbiome communication is still mostly rudimentary and plant–microbiome interactions varies between crops species (and even cultivars), between individual members of the microbiome and with environmental conditions. In each individual case, many aspects of the plant–microorganisms relationship should be thoroughly scrutinized. In this article we summarize some of the existing plant–microbiome engineering studies and point out potential avenues for further research.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants.
- Record: found
- Abstract: not found
- Article: not found
Regulation and function of root exudates
- Record: found
- Abstract: found
- Article: not found