- Record: found
- Abstract: found
- Article: found
Hiding on jagged karst pinnacles: A new microendemic genus and species of a limestone-dwelling agamid lizard (Squamata: Agamidae: Draconinae) from Khammouan Province, Laos
Read this article at
Abstract
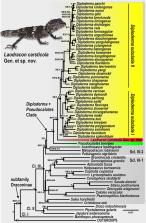
We describe a unique new species and genus of agamid lizard from the karstic massifs of Khammouan Province, central Laos. Laodracon carsticola Gen. et sp. nov. is an elusive medium-sized lizard (maximum snout-vent length 101 mm) specifically adapted to life on limestone rocks and pinnacles. To assess the phylogenetic position of the new genus amongst other agamids, we generated DNA sequences from two mitochondrial gene fragments (16S rRNA and ND2) and three nuclear loci ( BDNF, RAG1 and c-mos), with a final alignment comprising 7 418 base pairs for 64 agamid species. Phylogenetic analyses unambiguously place the new genus in the mainland Asia subfamily Draconinae, where it forms a clade sister to the genus Diploderma from East Asia and the northern part of Southeast Asia. Morphologically, the new genus is distinguished from all other genera in Draconinae by possessing a notably swollen tail base with enlarged scales on its dorsal and ventral surfaces. Our work provides further evidence that limestone regions of Indochina represent unique “arks of biodiversity” and harbor numerous relict lineages. To date, Laodracon carsticola Gen. et sp. nov. is known from only two adult male specimens and its distribution seems to be restricted to a narrow limestone massif on the border of Khammouan and Bolikhamxai provinces of Laos. Additional studies are required to understand its life history, distribution, and conservation status.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.

- Record: found
- Abstract: found
- Article: found
IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies
- Record: found
- Abstract: found
- Article: not found