- Record: found
- Abstract: found
- Article: found
Identification of Targets from LRRK2 Rescue Phenotypes
Read this article at
Abstract
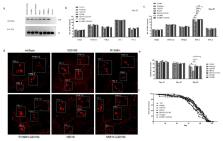
Parkinson’s disease (PD) is an age-dependent neurodegenerative condition. Leucine-rich repeat kinase 2 (LRRK2) mutations are the most frequent cause of sporadic and autosomal dominant PD. The exact role of LRRK2 protective variants (R1398H, N551K) together with a pathogenic mutant (G2019S) in aging and neurodegeneration is unknown. We generated the following myc-tagged UAS-LRRK2 transgenic Drosophila: LRRK2 (WT), N551K, R1398H, G2019S single allele, and double-mutants (N551K/G2019S or R1398H/G2019S). The protective variants alone were able to suppress the phenotypic effects caused by the pathogenic LRRK2 mutation. Next, we conducted RNA-sequencing using mRNA isolated from dopaminergic neurons of these different groups of transgenic Drosophila. Using pathway enrichment analysis, we identified the top 10 modules ( p < 0.05), with “LRRK2 in neurons in Parkinson’s disease” among the candidates. Further dissection of this pathway identified the most significantly modulated gene nodes such as eEF1A2, ACTB, eEF1A, and actin cytoskeleton reorganization. The induction of the pathway was successfully restored by the R1398H protective variant and R1398H-G2019S or N551K-G2019S rescue experiments. The oxidoreductase family of genes was also active in the pathogenic mutant and restored in protective and rescue variants. In summary, we provide in vivo evidence supporting the neuroprotective effects of LRRK2 variants. RNA sequencing of dopaminergic neurons identified upregulation of specific gene pathways in the Drosophila carrying the pathogenic variant, and this was restored in the rescue phenotypes. Using protective gene variants, our study identifies potential new targets and provides proof of principle of a new therapeutic approach that will further our understanding of aging and neurodegeneration in PD.
Related collections
Most cited references64
- Record: found
- Abstract: found
- Article: not found
Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks.
- Record: found
- Abstract: found
- Article: not found
TopHat: discovering splice junctions with RNA-Seq
- Record: found
- Abstract: found
- Article: not found