- Record: found
- Abstract: found
- Article: found
Deep amoA amplicon sequencing reveals community partitioning within ammonia-oxidizing bacteria in the environmentally dynamic estuary of the River Elbe
Read this article at
Abstract
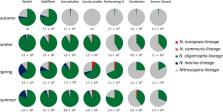
The community composition of betaproteobacterial ammonia-oxidizing bacteria (ß-AOB) in the River Elbe Estuary was investigated by high throughput sequencing of ammonia monooxygenase subunit A gene ( amoA) amplicons. In the course of the seasons surface sediment samples from seven sites along the longitudinal profile of the upper Estuary of the Elbe were investigated. We observed striking shifts of the ß-AOB community composition according to space and time. Members of the Nitrosomonas oligotropha-lineage and the genus Nitrosospira were found to be the dominant ß-AOB within the river transect, investigated. However, continuous shifts of balance between members of both lineages along the longitudinal profile were determined. A noticeable feature was a substantial increase of proportion of Nitrosospira-like sequences in autumn and of sequences affiliated with the Nitrosomonas marina-lineage at downstream sites in spring and summer. Slightly raised relative abundances of sequences affiliated with the Nitrosomonas europaea/Nitrosomonas mobilis-lineage and the Nitrosomonas communis-lineage were found at sampling sites located in the port of Hamburg. Comparisons between environmental parameters and AOB-lineage (ecotype) composition revealed promising clues that processes happening in the fluvial to marine transition zone of the Elbe estuary are reflected by shifts in the relative proportion of ammonia monooxygenase sequence abundance, and hence, we propose ß-AOB as appropriate indicators for environmental dynamics and the ecological condition of the Elbe Estuary.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
Complete nitrification by Nitrospira bacteria
- Record: found
- Abstract: found
- Article: not found