- Record: found
- Abstract: found
- Article: found
A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood–brain barrier in ddY, FVB, and C57BL/6J mice
Read this article at
Abstract
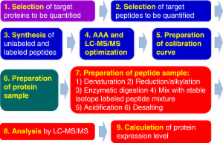
Proteomics has opened a new horizon in biological sciences. Global proteomic analysis is a promising technology for the discovery of thousands of proteins, post-translational modifications, polymorphisms, and molecular interactions in a variety of biological systems. The activities and roles of the identified proteins must also be elucidated, but this is complicated by the inability of conventional proteomic methods to yield quantitative information for protein expression. Thus, a variety of biological systems remain “black boxes”. Quantitative targeted absolute proteomics (QTAP) enables the determination of absolute expression levels (mol) of any target protein, including low-abundance functional proteins, such as transporters and receptors. Therefore, QTAP will be useful for understanding the activities and roles of individual proteins and their differences, including normal/disease, human/animal, or in vitro/ in vivo. Here, we describe the study protocols and precautions for QTAP experiments including in silico target peptide selection, determination of peptide concentration by amino acid analysis, setup of selected/multiple reaction monitoring (SRM/MRM) analysis in liquid chromatography–tandem mass spectrometry, preparation of protein samples (brain capillaries and plasma membrane fractions) followed by the preparation of peptide samples, simultaneous absolute quantification of target proteins by SRM/MRM analysis, data analysis, and troubleshooting. An application of QTAP in biological sciences was introduced that utilizes data from inter-strain differences in the protein expression levels of transporters, receptors, tight junction proteins and marker proteins at the blood–brain barrier in ddY, FVB, and C57BL/6J mice. Among 18 molecules, 13 (abcb1a/mdr1a/P-gp, abcc4/mrp4, abcg2/bcrp, slc2a1/glut1, slc7a5/lat1, slc16a1/mct1, slc22a8/oat3, insr, lrp1, tfr1, claudin-5, Na +/K +-ATPase, and γ-gtp) were detected in the isolated brain capillaries, and their protein expression levels were within a range of 0.637-101 fmol/μg protein. The largest difference in the levels between the three strains was 2.2-fold for 13 molecules, although bcrp and mct1 displayed statistically significant differences between C57BL/6J and the other strain(s). Highly sensitive simultaneous absolute quantification achieved by QTAP will increase the usefulness of proteomics in biological sciences and is expected to advance the new research field of pharmacoproteomics (PPx).
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: not found
Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins.
- Record: found
- Abstract: found
- Article: not found
Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria.
- Record: found
- Abstract: found
- Article: not found