- Record: found
- Abstract: found
- Article: found
Down-Regulation of ECRG4, a Candidate Tumor Suppressor Gene, in Human Breast Cancer
Read this article at
Abstract
Introduction
ECRG4/C2ORF40 is a potential tumor suppressor gene (TSG) recently identified in esophageal carcinoma. Its expression, gene copy number and prognostic value have never been explored in breast cancer.
Methods
Using DNA microarray and array-based comparative genomic hybridization (aCGH), we examined ECRG4 mRNA expression and copy number alterations in 353 invasive breast cancer samples and normal breast (NB) samples. A meta-analysis was done on a large public retrospective gene expression dataset (n = 1,387) in search of correlations between ECRG4 expression and histo-clinical features including survival.
Results
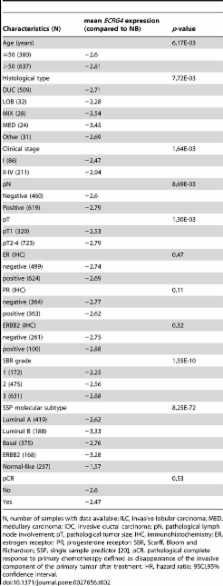
ECRG4 was underexpressed in 94.3% of cancers when compared to NB. aCGH data revealed ECRG4 loss in 18% of tumors, suggesting that DNA loss is not the main mechanism of underexpression. Meta-analysis showed that ECRG4 expression was significantly higher in tumors displaying earlier stage, smaller size, negative axillary lymph node status, lower grade, and normal-like subtype. Higher expression was also associated with disease-free survival (DFS; HR = 0.84 [0.76–0.92], p = 0.0002) and overall survival (OS; HR = 0.72 [0.63–0.83], p = 5.0E-06). In multivariate analysis including the other histo-clinical prognostic features, ECRG4 expression remained the only prognostic factor for DFS and OS.
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts
- Record: found
- Abstract: found
- Article: not found
Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer.
- Record: found
- Abstract: found
- Article: not found