- Record: found
- Abstract: found
- Article: found
Effect of Environmental Factors and an Emerging Parasitic Disease on Gut Microbiome of Wild Salmonid Fish

Read this article at
Abstract
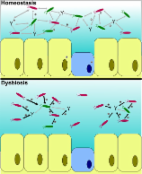
Cohabiting microorganisms play diverse and important roles in the biology of multicellular hosts, but their diversity and interactions with abiotic and biotic factors remain largely unsurveyed. Nevertheless, it is becoming increasingly clear that many properties of host phenotypes reflect contributions from the associated microbiome. We focus on a question of how parasites, the host genetic background, and abiotic factors influence the microbiome in salmonid hosts by using a host-parasite model consisting of wild brown trout ( Salmo trutta) and the myxozoan Tetracapsuloides bryosalmonae, which causes widely distributed proliferative kidney disease. We show that parasite infection increases the frequency of bacteria from the surrounding river water community, reflecting impaired homeostasis in the fish gut. Our results also demonstrate the importance of abiotic environmental factors and host size in the assemblage of the gut microbiome of fish and the relative insignificance of the host genotype and gender.
ABSTRACT
The gastrointestinal tract (GIT) of fish supports a dynamic microbial ecosystem that is intimately linked to host nutrient acquisition, epithelial development, immune system priming, and disease prevention, and we are far from understanding the complex interactions among parasites, symbiotic gut bacteria, and host fitness. Here, we analyzed the effects of environmental factors and parasitic burdens on the microbial composition and diversity within the GIT of the brown trout ( Salmo trutta). We focused on the emerging dangerous salmonid myxozoan parasite Tetracapsuloides bryosalmonae, which causes proliferative kidney disease in salmonid fish, to demonstrate the potential role of GIT micobiomes in the modulation of host-parasite relationships. The microbial diversity in the GIT displayed clear clustering according to the river of origin, while considerable variation was also found among fish from the same river. Environmental variables such as oxygen concentration, water temperature, and river morphometry strongly associated with both the river microbial community and the GIT microbiome, supporting the role of the environment in microbial assemblage and the relative insignificance of the host genotype and gender. Contrary to expectations, the parasite load exhibited a significant positive relationship with the richness of the GIT microbiome. Many operational taxonomic units (OTUs; n = 202) are more abundant in T. bryosalmonae-infected fish, suggesting that brown trout with large parasite burdens are prone to lose their GIT microbiome homeostasis. The OTUs with the strongest increase in infected trout are mostly nonpathogenic aquatic, anaerobic sediment/sludge, or ruminant bacteria. Our results underscore the significance of the interactions among parasitic disease, abiotic factors, and the GIT microbiome in disease etiology.
IMPORTANCE Cohabiting microorganisms play diverse and important roles in the biology of multicellular hosts, but their diversity and interactions with abiotic and biotic factors remain largely unsurveyed. Nevertheless, it is becoming increasingly clear that many properties of host phenotypes reflect contributions from the associated microbiome. We focus on a question of how parasites, the host genetic background, and abiotic factors influence the microbiome in salmonid hosts by using a host-parasite model consisting of wild brown trout ( Salmo trutta) and the myxozoan Tetracapsuloides bryosalmonae, which causes widely distributed proliferative kidney disease. We show that parasite infection increases the frequency of bacteria from the surrounding river water community, reflecting impaired homeostasis in the fish gut. Our results also demonstrate the importance of abiotic environmental factors and host size in the assemblage of the gut microbiome of fish and the relative insignificance of the host genotype and gender.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
Microbiota-mediated colonization resistance against intestinal pathogens.
- Record: found
- Abstract: found
- Article: not found