- Record: found
- Abstract: found
- Article: found
Rescue of HIV-1 Release by Targeting Widely Divergent NEDD4-Type Ubiquitin Ligases and Isolated Catalytic HECT Domains to Gag
Read this article at
Abstract
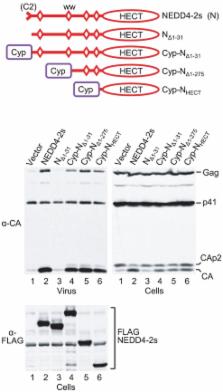
Retroviruses engage the ESCRT pathway through late assembly (L) domains in Gag to promote virus release. HIV-1 uses a PTAP motif as its primary L domain, which interacts with the ESCRT-I component Tsg101. In contrast, certain other retroviruses primarily use PPxY-type L domains, which constitute ligands for NEDD4-type ubiquitin ligases. Surprisingly, although HIV-1 Gag lacks PPxY motifs, the release of HIV-1 L domain mutants is potently enhanced by ectopic NEDD4-2s, a native isoform with a naturally truncated C2 domain that appears to account for the residual titer of L domain-defective HIV-1. The reason for the unique potency of the NEDD4-2s isoform has remained unclear. We now show that the naturally truncated C2 domain of NEDD4-2s functions as an autonomous Gag-targeting module that can be functionally replaced by the unrelated Gag-binding protein cyclophilin A (CypA). The residual C2 domain of NEDD4-2s was sufficient to transfer the ability to stimulate HIV-1 budding to other NEDD4 family members, including the yeast homologue Rsp5, and even to isolated catalytic HECT domains. The isolated catalytic domain of NEDD4-2s also efficiently promoted HIV-1 budding when targeted to Gag via CypA. We conclude that the regions typically required for substrate recognition by HECT ubiquitin ligases are all dispensable to stimulate HIV-1 release, implying that the relevant target for ubiquitination is Gag itself or can be recognized by divergent isolated HECT domains. However, the mere ability to ubiquitinate Gag was not sufficient to stimulate HIV-1 budding. Rather, our results indicate that the synthesis of K63-linked ubiquitin chains is critical for ubiquitin ligase-mediated virus release.
Author Summary
To promote its escape from cells, HIV-1 hijacks cellular budding machinery through so-called L domains in its structural Gag protein. However, HIV-1 lacks a type of L domain that recruits NEDD4 ubiquitin ligases, a family of cellular enzymes that attach one or more copies of a small protein called ubiquitin to other proteins. Surprisingly, one NEDD4 family member, which is known as NEDD4-2s and stands out because its membrane-binding domain is uniquely truncated, can nevertheless potently stimulate HIV-1 release. Our study reveals that NEDD4-2s can do this because its altered membrane-binding domain allows it to associate with HIV-1 Gag. Remarkably, when tagged with the altered membrane-binding domain of NEDD4-2s, even a distantly related yeast protein becomes capable of stimulating the release of HIV-1. We also show that only the portion of NEDD4-2s that acts as an enzyme is required when targeted to HIV-1 Gag in an alternative manner. Taken together, our findings indicate that it is not simply the ability to attach ubiquitin to Gag, but rather the ability to form a particular type of ubiquitin chain in the immediate vicinity of Gag, that is critical to stimulate virus release.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins.
- Record: found
- Abstract: found
- Article: not found
Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension.
- Record: found
- Abstract: found
- Article: not found