- Record: found
- Abstract: found
- Article: found
Data-Derived Modeling Characterizes Plasticity of MAPK Signaling in Melanoma
Read this article at
Abstract
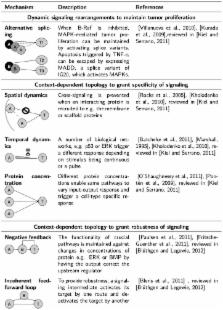
The majority of melanomas have been shown to harbor somatic mutations in the RAS-RAF-MEK-MAPK and PI3K-AKT pathways, which play a major role in regulation of proliferation and survival. The prevalence of these mutations makes these kinase signal transduction pathways an attractive target for cancer therapy. However, tumors have generally shown adaptive resistance to treatment. This adaptation is achieved in melanoma through its ability to undergo neovascularization, migration and rearrangement of signaling pathways. To understand the dynamic, nonlinear behavior of signaling pathways in cancer, several computational modeling approaches have been suggested. Most of those models require that the pathway topology remains constant over the entire observation period. However, changes in topology might underlie adaptive behavior to drug treatment. To study signaling rearrangements, here we present a new approach based on Fuzzy Logic (FL) that predicts changes in network architecture over time. This adaptive modeling approach was used to investigate pathway dynamics in a newly acquired experimental dataset describing total and phosphorylated protein signaling over four days in A375 melanoma cell line exposed to different kinase inhibitors. First, a generalized strategy was established to implement a parameter-reduced FL model encoding non-linear activity of a signaling network in response to perturbation. Next, a literature-based topology was generated and parameters of the FL model were derived from the full experimental dataset. Subsequently, the temporal evolution of model performance was evaluated by leaving time-defined data points out of training. Emerging discrepancies between model predictions and experimental data at specific time points allowed the characterization of potential network rearrangement. We demonstrate that this adaptive FL modeling approach helps to enhance our mechanistic understanding of the molecular plasticity of melanoma.
Author Summary
Signal transduction pathways can be described as static routes, transmitting extrinsic signals to the nucleus to induce a transcriptional response. In contrast to this reductionist view, the emerging paradigm is that signaling networks undergo dynamic crosstalk, both in disease and physiological conditions. To understand complex pathway behavior, it is necessary to develop methods to identify pathway interactions that are active as a consequence of stimuli and, importantly, to describe their evolution in time. To that end, we developed a method relying on prior knowledge networks in order to predict signaling crosstalk evolution, in response to perturbation and over time. The challenge we addressed was to establish a method dependent on information related to the topology of reported interactions, and not their mechanistic characteristics, and at the same time complex enough to reproduce the behavior of the signaling intermediates. The work presented here demonstrates that such an approach can be used to predict mechanisms that melanoma uses to rearrange its signaling and maintain its abnormal proliferation upon treatment.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation.
- Record: found
- Abstract: found
- Article: not found
Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K.
- Record: found
- Abstract: found
- Article: not found