- Record: found
- Abstract: found
- Article: found
Phylogenomic analyses of echinoid diversification prompt a re-evaluation of their fossil record
Read this article at
Abstract
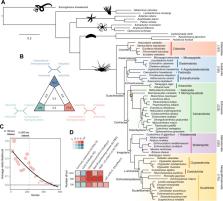
Echinoids are key components of modern marine ecosystems. Despite a remarkable fossil record, the emergence of their crown group is documented by few specimens of unclear affinities, rendering their early history uncertain. The origin of sand dollars, one of its most distinctive clades, is also unclear due to an unstable phylogenetic context. We employ 18 novel genomes and transcriptomes to build a phylogenomic dataset with a near-complete sampling of major lineages. With it, we revise the phylogeny and divergence times of echinoids, and place their history within the broader context of echinoderm evolution. We also introduce the concept of a chronospace – a multidimensional representation of node ages – and use it to explore methodological decisions involved in time calibrating phylogenies. We find the choice of clock model to have the strongest impact on divergence times, while the use of site-heterogeneous models and alternative node prior distributions show minimal effects. The choice of loci has an intermediate impact, affecting mostly deep Paleozoic nodes, for which clock-like genes recover dates more congruent with fossil evidence. Our results reveal that crown group echinoids originated in the Permian and diversified rapidly in the Triassic, despite the relative lack of fossil evidence for this early diversification. We also clarify the relationships between sand dollars and their close relatives and confidently date their origins to the Cretaceous, implying ghost ranges spanning approximately 50 million years, a remarkable discrepancy with their rich fossil record.
Related collections
Most cited references139

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found