- Record: found
- Abstract: found
- Article: found
Transient T-bet expression functionally specifies a distinct T follicular helper subset
Read this article at
Abstract
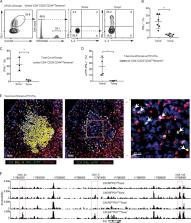
The mechanisms underlying the differentiation of T follicular helper (Tfh) cell subsets are poorly understood. Here, Fang et al. show that the NKG2D high Tfh cells in germinal centers with a history of T-bet expression represent the IFN-γ–producing Tfh subset.
Abstract
T follicular helper (Tfh) cells express transcription factor BCL-6 and cytokine IL-21. Mature Tfh cells are also capable of producing IFN-γ without expressing the Th1 transcription factor T-bet. Whether this IFN-γ–producing Tfh population represents a unique Tfh subset with a distinct differentiation pathway is poorly understood. By using T-bet fate–mapping mouse strains, we discovered that almost all the IFN-γ–producing Tfh cells have previously expressed T-bet and express high levels of NKG2D. DNase I hypersensitivity analysis indicated that the Ifng gene locus is partially accessible in this “ex–T-bet” population with a history of T-bet expression. Furthermore, multicolor tissue imaging revealed that the ex–T-bet Tfh cells found in germinal centers express IFN-γ in situ. Finally, we found that IFN-γ–expressing Tfh cells are absent in T-bet–deficient mice, but fully present in mice with T-bet deletion at late stages of T cell differentiation. Together, our findings demonstrate that transient expression of T-bet epigenetically imprints the Ifng locus for cytokine production in this Th1-like Tfh cell subset.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
High-resolution mapping and characterization of open chromatin across the genome.
- Record: found
- Abstract: found
- Article: not found
A clustering approach for identification of enriched domains from histone modification ChIP-Seq data.
- Record: found
- Abstract: found
- Article: not found