- Record: found
- Abstract: found
- Article: found
Effects of distinct drugs on gene transcription in an osteosarcoma cell line
Read this article at
Abstract
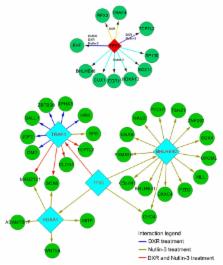
Osteosarcoma (OS) is a common cancerous bone tumor which has a detrimental impact on the lives of patients and their families. The present study aimed at investigating the underlying molecular mechanism of various drug treatments pertaining to OS, including dimethyl sulfoxide (DMSO), doxorubicin (DXP), Nutlin-3, actinomycin D (ActD) and etoposide (Eto). Microarray and p53 chromatin immunoprecipitation combined with sequencing (ChIP-seq) datasets of the OS cell line U2OS treated with distinct drugs were acquired from the Gene Expression Omnibus and differentially-expressed genes (DEGs) were screened for alignment analysis. The p53-binding target genes were identified and ChIP-seq and microarray gene expression data were combined to identify directly and indirectly targeted genes. A regulatory network of p53 was constructed with the acquired data. Finally, the Database for Annotation, Visualization and Integrated Discovery was interrogated for annotation of target genes. A total of 212 p53-binding peaks were obtained in the untreated group, whereas thousands of peaks were obtained in the treated groups. In total, ~1,000 target genes were identified in each of DXP, DMSO, Eto and ActD treatment groups, whereas the Nutlin-3 treatment group identified an increased number, with 5,458 target genes obtained. Several common DEGs including MDM2, TP53I3, RRM2B, FAS and SESN1 were targeted by all the drugs with the exception of DMSO. p53 regulated various genes including EHF, HOXA10 and BHLHE40 in the Nutlin-3 treatment group, whereas p53 regulated EHF, RFX3, TRAF40 and TCF7L2 in the DXR treatment group. The results of the present study indicate that p53 was able to directly regulate target genes including MDM2, TP53I3 and RRM2B or indirectly regulate numerous further genes through several hub genes including EHF and RFX through various drug treatments in U2OS cells. Furthermore, p53 regulated distinct molecular processes in various drug treatments.
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes.
- Record: found
- Abstract: found
- Article: not found
Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities.
- Record: found
- Abstract: found
- Article: not found