- Record: found
- Abstract: found
- Article: found
Diacylglycerol-evoked activation of PKC and PKD isoforms in regulation of glucose and lipid metabolism: a review
Read this article at
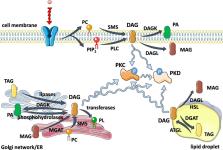
Abstract
Protein kinase C (PKC) and Protein kinase D (PKD) isoforms can sense diacylglycerol (DAG) generated in the different cellular compartments in various physiological processes. DAG accumulates in multiple organs of the obese subjects, which leads to the disruption of metabolic homeostasis and the development of diabetes as well as associated diseases. Multiple studies proved that aberrant activation of PKCs and PKDs contributes to the development of metabolic diseases. DAG-sensing PKC and PKD isoforms play a crucial role in the regulation of metabolic homeostasis and therefore might serve as targets for the treatment of metabolic disorders such as obesity and diabetes.
Related collections
Most cited references171
- Record: found
- Abstract: found
- Article: not found
Signaling pathways in skeletal muscle remodeling.
- Record: found
- Abstract: found
- Article: not found
Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation.
- Record: found
- Abstract: found
- Article: not found
