- Record: found
- Abstract: found
- Article: found
The terminal enzymes of (bacterio)chlorophyll biosynthesis
Read this article at
Abstract
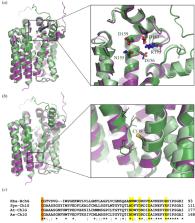
(Bacterio)chlorophylls are modified tetrapyrroles that are used by phototrophic organisms to harvest solar energy, powering the metabolic processes that sustain most of the life on Earth. Biosynthesis of these pigments involves enzymatic modification of the side chains and oxidation state of a porphyrin precursor, modifications that differ by species and alter the absorption properties of the pigments. (Bacterio)chlorophylls are coordinated by proteins that form macromolecular assemblies to absorb light and transfer excitation energy to a special pair of redox-active (bacterio)chlorophyll molecules in the photosynthetic reaction centre. Assembly of these pigment–protein complexes is aided by an isoprenoid moiety esterified to the (bacterio)chlorin macrocycle, which anchors and stabilizes the pigments within their protein scaffolds. The reduction of the isoprenoid ‘tail’ and its addition to the macrocycle are the final stages in (bacterio)chlorophyll biosynthesis and are catalysed by two enzymes, geranylgeranyl reductase and (bacterio)chlorophyll synthase. These enzymes work in conjunction with photosynthetic complex assembly factors and the membrane biogenesis machinery to synchronize delivery of the pigments to the proteins that coordinate them. In this review, we summarize current understanding of the catalytic mechanism, substrate recognition and regulation of these crucial enzymes and their involvement in thylakoid biogenesis and photosystem repair in oxygenic phototrophs.
Related collections
Most cited references211

- Record: found
- Abstract: found
- Article: found
Highly accurate protein structure prediction with AlphaFold
- Record: found
- Abstract: found
- Article: not found
Reactive oxygen species: metabolism, oxidative stress, and signal transduction.
- Record: found
- Abstract: found
- Article: found