- Record: found
- Abstract: found
- Article: found
Toxoplasma gondii suppresses proliferation and migration of breast cancer cells by regulating their transcriptome
Read this article at
Abstract
Background
Breast cancer is the most common cancer in women worldwide. Toxoplasma gondii ( T. gondii) has shown anticancer activity in breast cancer mouse models, and exerted beneficial effect on the survival of breast cancer patients, but the mechanism was unclear.
Methods
The effect of tachyzoites of T. gondii (RH and ME49 strains) on human breast cancer cells (MCF-7 and MDA-MB-231 cells) proliferation and migration was assessed using cell growth curve and wound healing assays. Dual RNA-seq was performed for T. gondii-infected and non-infected cells to determine the differentially expressed genes (DEGs). Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Protein–Protein Interaction Networks analysis (PPI) were performed to explore the related signaling pathway and hub genes. Hub genes were validated using the Kaplan–Meier plotter database, and Pathogen Host Interaction (PHI-base) database. The results were verified by qRT-PCR.
Results
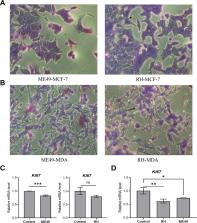
The tachyzoites of T. gondii decreased the expression of Ki67 and increased the expression of E-cadherin, resulting in suppressing the proliferation and migration of infected human breast cancer cells. The inhibitory effect of T. gondii on breast cancer cells showed a significant dose–response relationship. Compared with the control group, 2321 genes were transcriptionally regulated in MCF-7 cells infected with T. gondii, while 169 genes were transcriptionally regulated in infected MDA-MB-231 cells. Among these genes, 698 genes in infected MCF-7 cells and 67 genes in infected MDA-MB-231 cells were validated by the publicly available database. GO and KEGG analyses suggested that several pathways were involved in anticancer function of T. gondii, such as ribosome, interleukin-17 signaling, coronavirus disease pathway, and breast cancer pathway. BRCA1, MYC and IL-6 were identified as the top three hub genes in infected-breast cancer cells based on the connectivity of PPI analysis. In addition, after interacting with breast cancer cells, the expression of ROP16 and ROP18 in T. gondii increased, while the expression of crt, TgIST, GRA15, GRA24 and MIC13 decreased.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found