- Record: found
- Abstract: found
- Article: found
Draft Genome Sequence of Enterobacter asburiae UFMG-H9, Isolated from Urine from a Healthy Bovine Heifer (Gyr Breed)
brief-report
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
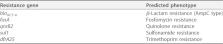
Enterobacter asburiae is part of the Enterobacter cloacae complex, related to nosocomial opportunistic infections in humans. Here, we report the draft genome of E. asburiae strain UFMG-H9, an isolate from urine from a healthy Gyr heifer.
ABSTRACT
Enterobacter asburiae is part of the Enterobacter cloacae complex, related to nosocomial opportunistic infections in humans. Here, we report the draft genome of E. asburiae strain UFMG-H9, an isolate from urine from a healthy Gyr heifer.
Related collections
Most cited references4
- Record: found
- Abstract: found
- Article: not found
Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA.
J. R. Marchesi, T Sato, A. Weightman … (1998)

- Record: found
- Abstract: found
- Article: not found