- Record: found
- Abstract: found
- Article: not found
Significant inhibition of re-emerged and emerging swine enteric coronavirus in vitro using the multiple shRNA expression vector
Abstract
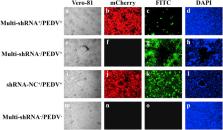
Swine enteric coronaviruses (SECoVs), including porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV), and porcine deltacoronavirus (PDCoV) have emerged and been prevalent in pig populations in China for the last several years. However, current traditional inactivated and attenuated PEDV vaccines are of limited efficacy against circulating PEDV variants, and there are no commercial vaccines for prevention of PDCoV and SADS-CoV. RNA interference (RNAi) is a powerful tool in therapeutic applications to inhibit viral replication in vitro. In this study, we developed a small interfering RNA generation system that expressed two different short hairpin RNAs (shRNAs) targeting the M gene of PEDV and SADS-CoV and the N gene of PDCoV, respectively. Our results demonstrated that simultaneous expression of these specific shRNA molecules inhibited expression of PEDV M gene, SADS-CoV M gene, and PDCoV N gene RNA by 99.7%, 99.4%, and 98.8%, respectively, in infected cell cultures. In addition, shRNA molecules significantly restricted the expression of M and N protein, and impaired the replication of PEDV, SADS-CoV, and PDCoV simultaneously. Taken together, this shRNAs expression system not only is proved to be a novel approach for studying functions of various genes synchronously, but also developed to test aspects of a potential therapeutic option for treatment and prevention of multiple SECoV infections.
Highlights
-
•
Two potential targets of antiviral molecules for the treatment of three swine enteric coronaviruses replication are tested.
-
•
The multiple-shRNA expression vector was constructed to effectively inhibit PEDV, SADS-CoV, and PDCoV.
-
•
A shRNA-based multiple-resistance antiviral strategy, significantly affecting viral replication, was evaluated in vitro.
Related collections
Most cited references39

- Record: found
- Abstract: found
- Article: found
Origin, Evolution, and Virulence of Porcine Deltacoronaviruses in the United States
- Record: found
- Abstract: found
- Article: found
New Variants of Porcine Epidemic Diarrhea Virus, China, 2011
- Record: found
- Abstract: found
- Article: not found
