- Record: found
- Abstract: found
- Article: found
Dimerization facilitates the conformational transitions for bacterial phosphotransferase enzyme I autophosphorylation in an allosteric manner
Read this article at
Abstract
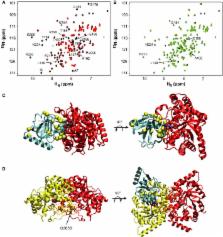
The bacterial phosphotransferase system is central to sugar uptake and phosphorylation. Enzyme I ( EI), the first enzyme of the system, autophosphorylates as a dimer using phosphoenolpyruvate ( PEP), but it is not clearly understood how dimerization activates the enzyme activity. Here, we show that EI dimerization is important for proper conformational transitions and the domain association required for the autophosphorylation. EI(G356S) with reduced dimerization affinity and lower autophosphorylation activity revealed that significantly hindered conformational transitions are required for the phosphoryl transfer reaction. The G356S mutation does not change the binding affinity for PEP, but perturbs the domain association accompanying large interdomain motions that bring the active site His189 close to PEP. The interface for the domain association is separate from the dimerization interface, demonstrating that dimerization can prime the conformational change in an allosteric manner.
Related collections
Most cited references22
- Record: found
- Abstract: found
- Article: not found
The changing landscape of protein allostery.
- Record: found
- Abstract: found
- Article: not found
Principles of Allosteric Interactions in Cell Signaling
- Record: found
- Abstract: found
- Article: not found