- Record: found
- Abstract: found
- Article: found
Partitioning of diet between species and life history stages of sympatric and cryptic snappers (Lutjanidae) based on DNA metabarcoding
Read this article at
Abstract
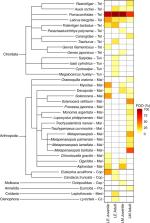
Lutjanus erythropterus and L. malabaricus are sympatric, sister taxa that are important to fisheries throughout the Indo-Pacific. Their juveniles are morphologically indistinguishable (i.e. cryptic). A DNA metabarcoding dietary study was undertaken to assess the diet composition and partitioning between the juvenile and adult life history stages of these two lutjanids. Major prey taxa were comprised of teleosts and crustaceans for all groups except adult L. erythropterus, which instead consumed soft bodied invertebrates (e.g. tunicates, comb jellies and medusae) as well as teleosts, with crustaceans being notably absent. Diet composition was significantly different among life history stages and species, which may be associated with niche habitat partitioning or differences in mouth morphology within adult life stages. This study provides the first evidence of diet partitioning between cryptic juveniles of overlapping lutjanid species, thus providing new insights into the ecological interactions, habitat associations, and the specialised adaptations required for the coexistence of closely related species. This study has improved our understanding of the differential contributions of the juvenile and adult diets of these sympatric species within food webs. The diet partitioning reported in this study was only revealed by the taxonomic resolution provided by the DNA metabarcoding approach and highlights the potential utility of this method to refine the dietary components of reef fishes more generally.
Related collections
Most cited references45

- Record: found
- Abstract: found
- Article: found
Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data?
- Record: found
- Abstract: found
- Article: not found
GenBank
- Record: found
- Abstract: found
- Article: not found