- Record: found
- Abstract: found
- Article: found
Germline variation in BRCA1/ 2 is highly ethnic‐specific: Evidence from over 30,000 Chinese hereditary breast and ovarian cancer patients
Read this article at
Abstract
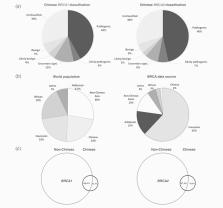
BRCA1 and BRCA2 play essential roles in maintaining the genome stability. Pathogenic germline mutations in these two genes disrupt their function, lead to genome instability and increase the risk of developing breast and ovarian cancers. BRCA mutations have been extensively screened in Caucasian populations, and the resulting information are used globally as the standard reference in clinical diagnosis, treatment and prevention of BRCA‐related cancers. Recent studies suggest that BRCA mutations can be ethnic‐specific, raising the question whether a Caucasian‐based BRCA mutation information can be used as a universal standard worldwide, or whether an ethnicity‐based BRCA mutation information system need to be developed for the corresponding ethnic populations. In this study, we used Chinese population as a model to test ethnicity‐specific BRCA mutations considering that China has one of the latest numbers of breast cancer patients therefore BRCA mutation carriers. Through comprehensive data mining, standardization and annotation, we collected 1,088 distinct BRCA variants derived from over 30,000 Chinese individuals, one of the largest BRCA data set from a non‐Caucasian population covering nearly all known BRCA variants in the Chinese population ( https://dbBRCA-Chinese.fhs.umac.mo). Using this data, we performed multi‐layered analyses to determine the similarities and differences of BRCA variation between Chinese and non‐Chinese ethnic populations. The results show the substantial differences of BRCA data between Chinese and non‐Chinese ethnicities. Our study indicates that the current Caucasian population‐based BRCA data is not adequate to represent the BRCA status in non‐Caucasian populations. Therefore, ethnic‐based BRCA standards need to be established to serve for the non‐Caucasian populations.
Abstract
What's new?
Currently, Caucasian population‐based BRCA mutation data are used worldwide as the standard reference for diagnosis, treatment, and prevention of BRCA‐associated cancers. Recent studies however suggest that BRCA variation can be ethnic specific. Here, the authors carried out a comprehensive comparison of BRCA mutation data between the Chinese and worldwide non‐Chinese populations and found substantial differences. The study suggests that BRCA mutations are highly ethnic specific and that the current Caucasian population‐based BRCA data is not adequate to represent the BRCA status in non‐Caucasian populations. Developing new standard references using ethnic‐based BRCA mutation data is needed to better serve non‐Caucasian ethnic populations.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1.
- Record: found
- Abstract: found
- Article: not found
dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs.
- Record: found
- Abstract: found
- Article: not found