- Record: found
- Abstract: found
- Article: found
A Family of Viral Satellites Manipulates Invading Virus Gene Expression and Can Affect Cholera Toxin Mobilization
Read this article at
Abstract
Viral satellites are found in all domains of life and can have profound fitness effects on both the viruses they parasitize and the cells they reside in. In this study, we have acquired the first RNA sequencing (RNA-seq) transcriptomes of viral satellites outside plants, as well as the transcriptome of the phage ICP1, a predominant predator of pandemic Vibrio cholerae. Capsid downregulation, previously observed in an unrelated phage satellite, is conserved among phage inducible chromosomal island- like elements (PLEs), suggesting that viral satellites are under strong selective pressure to reduce the capsid expression of their larger host viruses. Despite conserved manipulation of capsid expression, PLEs exhibit divergent effects on CTXΦ transcription and mobility. Our results demonstrate that PLEs can influence both their hosts’ resistance to phage and the mobility of virulence-encoding elements, suggesting that PLEs can play a substantial role in shaping Vibrio cholerae evolution.
ABSTRACT
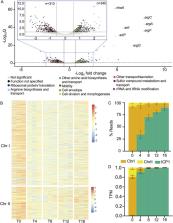
Many viruses possess temporally unfolding gene expression patterns aimed at subverting host defenses, commandeering host metabolism, and ultimately producing a large number of progeny virions. High-throughput omics tools, such as RNA sequencing (RNA-seq), have dramatically enhanced the resolution of expression patterns during infection. Less studied have been viral satellites, mobile genomes that parasitize viruses. By performing RNA-seq on infection time courses, we have obtained the first time-resolved transcriptomes for bacteriophage satellites during lytic infection. Specifically, we have acquired transcriptomes for the lytic Vibrio cholerae phage ICP1 and all five known variants of ICP1’s parasite, the phage inducible chromosomal island- like elements (PLEs). PLEs rely on ICP1 for both DNA replication and mobilization and abolish production of ICP1 progeny in infected cells. We investigated PLEs’ impact on ICP1 gene expression and found that PLEs did not broadly restrict or reduce ICP1 gene expression. A major exception occurred in ICP1’s capsid morphogenesis operon, which was downregulated by each of the PLE variants. Surprisingly, PLEs were also found to alter the gene expression of CTXΦ, the integrative phage that encodes cholera toxin and is necessary for virulence of toxigenic V. cholerae. One PLE, PLE1, upregulated CTXΦ genes involved in replication and integration and boosted CTXΦ mobility following induction of the SOS response.
IMPORTANCE Viral satellites are found in all domains of life and can have profound fitness effects on both the viruses they parasitize and the cells they reside in. In this study, we have acquired the first RNA sequencing (RNA-seq) transcriptomes of viral satellites outside plants, as well as the transcriptome of the phage ICP1, a predominant predator of pandemic Vibrio cholerae. Capsid downregulation, previously observed in an unrelated phage satellite, is conserved among phage inducible chromosomal island- like elements (PLEs), suggesting that viral satellites are under strong selective pressure to reduce the capsid expression of their larger host viruses. Despite conserved manipulation of capsid expression, PLEs exhibit divergent effects on CTXΦ transcription and mobility. Our results demonstrate that PLEs can influence both their hosts’ resistance to phage and the mobility of virulence-encoding elements, suggesting that PLEs can play a substantial role in shaping Vibrio cholerae evolution.
Related collections
Most cited references72

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: found
- Article: not found