- Record: found
- Abstract: found
- Article: found
Recombinant polypeptide production in E. coli: towards a rational approach to improve the yields of functional proteins
Read this article at
Abstract
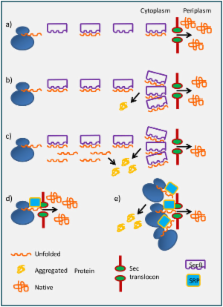
The development of complementary technologies enabled the successful production of recombinant polypeptides in bacteria and opened to biology researchers new avenues as obtaining suitable amounts of proteins necessary for their experimental work became easy, fast, and inexpensive. Nevertheless, the recombinant approach remained somehow unpredictable, since many constructs resisted to apparent production or accumulated as aggregates.
Several factors and physical/chemical conditions that could improve the accumulation of native-like protein were identified. At the same time, it was acknowledged that the outcome of most of them was erratic and that almost any protein required its own specific optimized set of conditions to achieve its correct folding. The attempt to understand the critical points specific for recombinant protein production missed the goal of setting universally useful protocols, but contributed to the increase of the rate of success by proposing always new empiric combinations.
Nevertheless, the results published in the recent literature allow for a better comprehension of some key mechanisms controlling protein production in E. coli and could enable the elaboration of rational methodologies for improving the quantitative and qualitative features of the produced polypeptides. This result will be achieved when the identification of the limiting step that impairs the accomplishment of the native folding for any single construct will become straightforward. This minireview will discuss how factors such as the expression rate, the folding machinery, and the secretion efficiency may impact the final protein yields.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels.
- Record: found
- Abstract: found
- Article: not found
Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli.
- Record: found
- Abstract: found
- Article: not found