- Record: found
- Abstract: found
- Article: found
Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways
Read this article at
Abstract
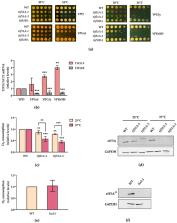
Translation elongation factor eIF5A binds to ribosomes to promote peptide bonds between problematic amino acids for the reaction like prolines. eIF5A is highly conserved and essential in eukaryotes, which usually contain two similar but differentially expressed paralogue genes. The human eIF5A-1 isoform is abundant and implicated in some cancer types; the eIF5A-2 isoform is absent in most cells but becomes overexpressed in many metastatic cancers. Several reports have connected eIF5A and mitochondria because it co-purifies with the organelle or its inhibition reduces respiration and mitochondrial enzyme levels. However, the mechanisms of eIF5A mitochondrial function, and whether eIF5A expression is regulated by the mitochondrial metabolism, are unknown. We analysed the expression of yeast eIF5A isoforms Tif51A and Tif51B under several metabolic conditions and in mutants. The depletion of Tif51A, but not Tif51B, compromised yeast growth under respiration and reduced oxygen consumption. Tif51A expression followed dual positive regulation: by high glucose through TORC1 signalling, like other translation factors, to promote growth and by low glucose or non-fermentative carbon sources through Snf1 and heme-dependent transcription factor Hap1 to promote respiration. Upon iron depletion, Tif51A was down-regulated and Tif51B up-regulated. Both were Hap1-dependent. Our results demonstrate eIF5A expression regulation by cellular metabolic status.
Related collections
Most cited references80
- Record: found
- Abstract: found
- Article: not found
Understanding the Warburg effect: the metabolic requirements of cell proliferation.
- Record: found
- Abstract: found
- Article: found
THE METABOLISM OF TUMORS IN THE BODY
- Record: found
- Abstract: found
- Article: not found