- Record: found
- Abstract: found
- Article: not found
UDP-galactose and Acetyl-CoA transporters as Plasmodium multidrug resistance genes
Abstract
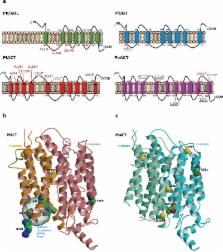
A molecular understanding of drug resistance mechanisms enables surveillance of the effectiveness of new antimicrobial therapies during development and deployment in the field. We used conventional drug resistance selection as well as a regime of limiting dilution at early stages of drug treatment to probe two antimalarial imidazolopiperazines, KAF156 and GNF179. The latter approach permits isolation of low-fitness mutants that might otherwise be out-competed during selection. Whole-genome sequencing of 24 independently-derived resistant P. falciparum clones revealed four parasites with mutations in the known cyclic amine resistance locus ( pfcarl), and a further 20 with mutations in two previously unreported P. falciparum drug resistance genes, an acetyl-CoA transporter ( pfact) and a UDP-galactose transporter ( pfugt). Mutations were validated both in vitro by CRISPR editing in P. falciparum, and in vivo by evolution of resistant P. berghei mutants. Both PfACT and PfUGT were localized to the endoplasmic reticulum by fluorescence microscopy. As mutations in pfact and pfugt conveyed resistance against additional unrelated chemical scaffolds, these genes are likely to be involved in broad mechanisms of antimalarial drug resistance.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery.
- Record: found
- Abstract: found
- Article: not found
Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening.
- Record: found
- Abstract: found
- Article: not found
