- Record: found
- Abstract: found
- Article: found
Novel CRISPR–Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives
Read this article at
Abstract
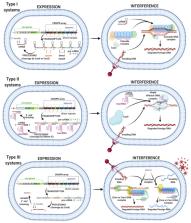
According to Darwin’s theory, endless evolution leads to a revolution. One such example is the Clustered Regularly Interspaced Palindromic Repeats (CRISPR)–Cas system, an adaptive immunity system in most archaea and many bacteria. Gene editing technology possesses a crucial potential to dramatically impact miscellaneous areas of life, and CRISPR–Cas represents the most suitable strategy. The system has ignited a revolution in the field of genetic engineering. The ease, precision, affordability of this system is akin to a Midas touch for researchers editing genomes. Undoubtedly, the applications of this system are endless. The CRISPR–Cas system is extensively employed in the treatment of infectious and genetic diseases, in metabolic disorders, in curing cancer, in developing sustainable methods for fuel production and chemicals, in improving the quality and quantity of food crops, and thus in catering to global food demands. Future applications of CRISPR–Cas will provide benefits for everyone and will save countless lives. The technology is evolving rapidly; therefore, an overview of continuous improvement is important. In this review, we aim to elucidate the current state of the CRISPR–Cas revolution in a tailor-made format from its discovery to exciting breakthroughs at the application level and further upcoming trends related to opportunities and challenges including ethical concerns.
Related collections
Most cited references268
- Record: found
- Abstract: found
- Article: not found
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity.
- Record: found
- Abstract: found
- Article: not found
CRISPR provides acquired resistance against viruses in prokaryotes.

- Record: found
- Abstract: found
- Article: found