- Record: found
- Abstract: found
- Article: found
Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis
Read this article at
Abstract
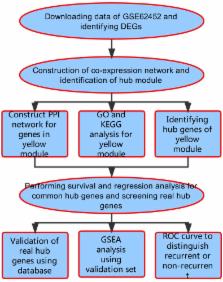
Since the five-year survival rate is less than 5%, pancreatic ductal adenocarcinoma (PDAC) remains the 4th cause of cancer-related death. Although PDAC has been repeatedly researched in recent years, it is still predicted to be the second leading cause of cancer death by year 2030. In our study, the differentially expressed genes in dataset GSE62452 were used to construct a co-expression network by WGCNA. The yellow module related to grade of PDAC was screened. Combined with co-expression network and PPI network, 36 candidates were screened. After survival and regression analysis by using GSE62452 and TCGA dataset, we identified 10 real hub genes ( CCNA2, CCNB1, CENPF, DLGAP5, KIF14, KIF23, NEK2, RACGAP1, TPX2 and UBE2C) tightly related to progression of PDAC. According to Oncomine database and The Human Protein Atlas (HPA), we found that all real hub genes were overexpressed in pancreatic carcinoma compared with normal tissues on transcriptional and translational level. ROC curve was plotted and AUC was calculated to distinguish recurrent and non-recurrent PDAC and every AUC of the real hub gene was greater than 0.5. Finally, functional enrichment analysis and gene set enrichment (GSEA) was performed and both of them showed the cell cycle played a vital role in PDAC.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma.
- Record: found
- Abstract: found
- Article: not found
The Role of Cholesterol in Cancer.
- Record: found
- Abstract: found
- Article: not found