- Record: found
- Abstract: found
- Article: found
Single-stranded heteroduplex intermediates in λ Red homologous recombination
Read this article at
Abstract
Background
The Red proteins of lambda phage mediate probably the simplest and most efficient homologous recombination reactions yet described. However the mechanism of dsDNA recombination remains undefined.
Results
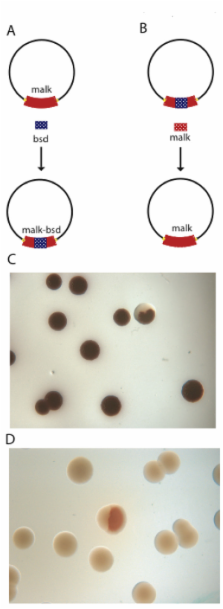
Here we show that the Red proteins can act via full length single stranded intermediates to establish single stranded heteroduplexes at the replication fork. We created asymmetrically digestible dsDNA substrates by exploiting the fact that Redα exonuclease activity requires a 5' phosphorylated end, or is blocked by phosphothioates. Using these substrates, we found that the most efficient configuration for dsDNA recombination occurred when the strand that can prime Okazaki-like synthesis contained both homology regions on the same ssDNA molecule. Furthermore, we show that Red recombination requires replication of the target molecule.
Conclusions
Hence we propose a new model for dsDNA recombination, termed 'beta' recombination, based on the formation of ssDNA heteroduplexes at the replication fork. Implications of the model were tested using (i) an in situ assay for recombination, which showed that recombination generated mixed wild type and recombinant colonies; and (ii) the predicted asymmetries of the homology arms, which showed that recombination is more sensitive to non-homologies attached to 5' than 3' ends. Whereas beta recombination can generate deletions in target BACs of at least 50 kb at about the same efficiency as small deletions, the converse event of insertion is very sensitive to increasing size. Insertions up to 3 kb are most efficiently achieved using beta recombination, however at greater sizes, an alternative Red-mediated mechanism(s) appears to be equally efficient. These findings define a new intermediate in homologous recombination, which also has practical implications for recombineering with the Red proteins.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Altering the genome by homologous recombination.
- Record: found
- Abstract: found
- Article: not found
The importance of repairing stalled replication forks.
- Record: found
- Abstract: found
- Article: not found