- Record: found
- Abstract: found
- Article: found
Microbial dynamics and metabolite changes in Chinese Rice Wine fermentation from sorghum with different tannin content
Read this article at
Abstract
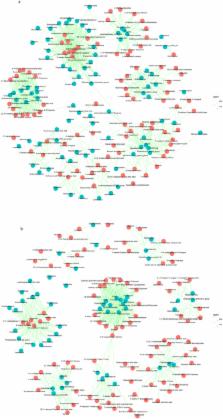
Chinese rice wine (CRW) is the oldest kind of wine in China and is mainly fermented by wheat Qu and yeast with rice, millet, etc. This gives CRW a unique quality, but the flavor components are complex. Its formation is related to microorganisms, but the link between CRW and microorganisms is poorly understood. Here, we used two kinds of sorghum (JZ22 and JB3, of which JZ22 has a higher tannin content) as the raw materials to brew and determined the structural and functional dynamics of the microbiota by metagenomics and flavor analyses. We detected 106 (JZ22) and 109 (JB3) volatile flavor compounds and 8 organic acids. By correlation analysis, we established 687 (JZ22) and 496 (JB3) correlations between the major flavor compounds and microbes. In JZ22, Blautia, Collinsella, Bifidobacterium, Faecalibacterium and Prevotella had the most correlations with flavor production. In JB3, the top 5 genera were Stenotrophomonas, Bdellovibrio, Solibacillus, Sulfuritalea and Achromobacter. In addition, more esters were detected, and more microorganisms correlated with ester generation in JZ22. This study provides a new idea for the micro ecological diversity of CRW fermented with sorghum. This is of significance for improving the quality and broadening the CRW varieties.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
FLASH: fast length adjustment of short reads to improve genome assemblies.
- Record: found
- Abstract: found
- Article: not found
454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity.
- Record: found
- Abstract: found
- Article: found