- Record: found
- Abstract: found
- Article: found
Trapped ion mobility spectrometry and PASEF enable in-depth lipidomics from minimal sample amounts
Read this article at
Abstract
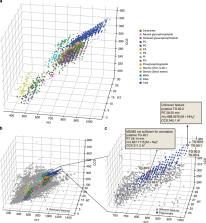
A comprehensive characterization of the lipidome from limited starting material remains very challenging. Here we report a high-sensitivity lipidomics workflow based on nanoflow liquid chromatography and trapped ion mobility spectrometry (TIMS). Taking advantage of parallel accumulation–serial fragmentation (PASEF), we fragment on average 15 precursors in each of 100 ms TIMS scans, while maintaining the full mobility resolution of co-eluting isomers. The acquisition speed of over 100 Hz allows us to obtain MS/MS spectra of the vast majority of isotope patterns. Analyzing 1 µL of human plasma, PASEF increases the number of identified lipids more than three times over standard TIMS-MS/MS, achieving attomole sensitivity. Building on high intra- and inter-laboratory precision and accuracy of TIMS collisional cross sections (CCS), we compile 1856 lipid CCS values from plasma, liver and cancer cells. Our study establishes PASEF in lipid analysis and paves the way for sensitive, ion mobility-enhanced lipidomics in four dimensions.
Abstract
Trapped ion mobility (TIMS)-mass spectrometry with parallel accumulation-serial fragmentation (PASEF) facilitates high-sensitivity proteomics experiments. Here, the authors expand TIMS and PASEF to small molecules and demonstrate fast and comprehensive lipidomics of low biological sample amounts.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics.

- Record: found
- Abstract: found
- Article: found
LMSD: LIPID MAPS structure database

- Record: found
- Abstract: found
- Article: found