- Record: found
- Abstract: found
- Article: found
Controversies Surrounding Segments and Parasegments in Onychophora: Insights from the Expression Patterns of Four “Segment Polarity Genes” in the Peripatopsid Euperipatoides rowelli
Read this article at
Abstract
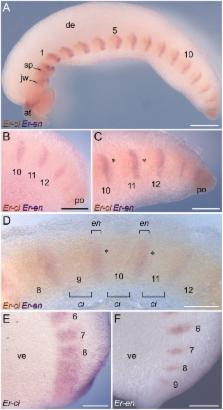
Arthropods typically show two types of segmentation: the embryonic parasegments and the adult segments that lie out of register with each other. Such a dual nature of body segmentation has not been described from Onychophora, one of the closest arthropod relatives. Hence, it is unclear whether onychophorans have segments, parasegments, or both, and which of these features was present in the last common ancestor of Onychophora and Arthropoda. To address this issue, we analysed the expression patterns of the “segment polarity genes” engrailed, cubitus interruptus, wingless and hedgehog in embryos of the onychophoran Euperipatoides rowelli. Our data revealed that these genes are expressed in repeated sets with a specific anterior-to-posterior order along the body in embryos of E. rowelli. In contrast to arthropods, the expression occurs after the segmental boundaries have formed. Moreover, the initial segmental furrow retains its position within the engrailed domain throughout development, whereas no new furrow is formed posterior to this domain. This suggests that no re-segmentation of the embryo occurs in E. rowelli. Irrespective of whether or not there is a morphological or genetic manifestation of parasegments in Onychophora, our data clearly show that parasegments, even if present, cannot be regarded as the initial metameric units of the onychophoran embryo, because the expression of key genes that define the parasegmental boundaries in arthropods occurs after the segmental boundaries have formed. This is in contrast to arthropods, in which parasegments rather than segments are the initial metameric units of the embryo. Our data further revealed that the expression patterns of “segment polarity genes” correspond to organogenesis rather than segment formation. This is in line with the concept of segmentation as a result of concerted evolution of individual periodic structures rather than with the interpretation of ‘segments’ as holistic units.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution.
- Record: found
- Abstract: found
- Article: not found
Hox genes and the evolution of the arthropod body plan.
- Record: found
- Abstract: found
- Article: not found