- Record: found
- Abstract: found
- Article: found
In silico Identification and Validation of a Linear and Naturally Immunogenic B-Cell Epitope of the Plasmodium vivax Malaria Vaccine Candidate Merozoite Surface Protein-9

Read this article at
Abstract
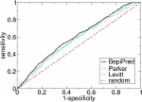
Synthetic peptide vaccines provide the advantages of safety, stability and low cost. The success of this approach is highly dependent on efficient epitope identification and synthetic strategies for efficacious delivery. In malaria, the Merozoite Surface Protein-9 of Plasmodium vivax (PvMSP9) has been considered a vaccine candidate based on the evidence that specific antibodies were able to inhibit merozoite invasion and recombinant proteins were highly immunogenic in mice and humans. However the identities of linear B-cell epitopes within PvMSP9 as targets of functional antibodies remain undefined. We used several publicly-available algorithms for in silico analyses and prediction of relevant B cell epitopes within PMSP9. We show that the tandem repeat sequence EAAPENAEPVHENA (PvMSP9 E795-A808) present at the C-terminal region is a promising target for antibodies, given its high combined score to be a linear epitope and located in a putative intrinsically unstructured region of the native protein. To confirm the predictive value of the computational approach, plasma samples from 545 naturally exposed individuals were screened for IgG reactivity against the recombinant PvMSP9-RIRII 729-972 and a synthetic peptide representing the predicted B cell epitope PvMSP9 E795-A808. 316 individuals (58%) were responders to the full repetitive region PvMSP9-RIRII, of which 177 (56%) also presented total IgG reactivity against the synthetic peptide, confirming it validity as a B cell epitope. The reactivity indexes of anti-PvMSP9-RIRII and anti-PvMSP9 E795-A808 antibodies were correlated. Interestingly, a potential role in the acquisition of protective immunity was associated with the linear epitope, since the IgG1 subclass against PvMSP9 E795-A808 was the prevalent subclass and this directly correlated with time elapsed since the last malaria episode; however this was not observed in the antibody responses against the full PvMSP9-RIRII. In conclusion, our findings identified and experimentally confirmed the potential of PvMSP9 E795-A808 as an immunogenic linear B cell epitope within the P. vivax malaria vaccine candidate PvMSP9 and support its inclusion in future subunit vaccines.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: found
Improved method for predicting linear B-cell epitopes
- Record: found
- Abstract: found
- Article: not found
Vivax malaria: neglected and not benign.
- Record: found
- Abstract: found
- Article: not found