- Record: found
- Abstract: found
- Article: not found
High-Resolution Structures and Orientations of Antimicrobial Peptides Piscidin 1 and Piscidin 3 in Fluid Bilayers Reveal Tilting, Kinking, and Bilayer Immersion

Read this article at
Abstract
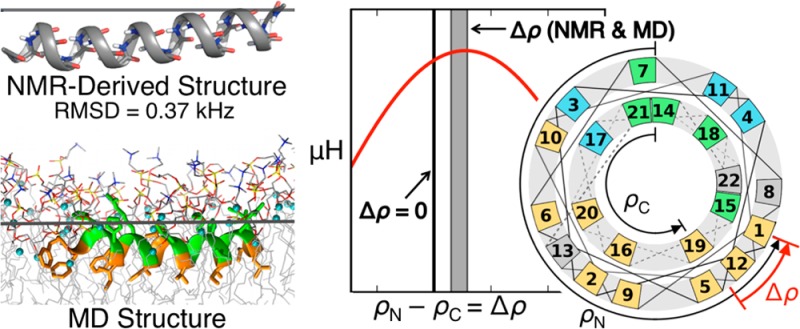
While antimicrobial peptides (AMPs) have been widely investigated as potential therapeutics, high-resolution structures obtained under biologically relevant conditions are lacking. Here, the high-resolution structures of the homologous 22-residue long AMPs piscidin 1 (p1) and piscidin 3 (p3) are determined in fluid-phase 3:1 phosphatidylcholine/phosphatidylglycerol (PC/PG) and 1:1 phosphatidylethanolamine/phosphatidylglycerol (PE/PG) bilayers to identify molecular features important for membrane destabilization in bacterial cell membrane mimics. Structural refinement of 1H– 15N dipolar couplings and 15N chemical shifts measured by oriented sample solid-state NMR and all-atom molecular dynamics (MD) simulations provide structural and orientational information of high precision and accuracy about these interfacially bound α-helical peptides. The tilt of the helical axis, τ, is between 83° and 93° with respect to the bilayer normal for all systems and analysis methods. The average azimuthal rotation, ρ, is 235°, which results in burial of hydrophobic residues in the bilayer. The refined NMR and MD structures reveal a slight kink at G13 that delineates two helical segments characterized by a small difference in their τ angles (<10°) and significant difference in their ρ angles (∼25°). Remarkably, the kink, at the end of a G(X) 4G motif highly conserved among members of the piscidin family, allows p1 and p3 to adopt ρ angles that maximize their hydrophobic moments. Two structural features differentiate the more potent p1 from p3: p1 has a larger ρ angle and less N-terminal fraying. The peptides have comparable depths of insertion in PC/PG, but p3 is 1.2 Å more deeply inserted than p1 in PE/PG. In contrast to the ideal α-helical structures typically assumed in mechanistic models of AMPs, p1 and p3 adopt disrupted α-helical backbones that correct for differences in the amphipathicity of their N- and C-ends, and their centers of mass lie ∼1.2–3.6 Å below the plane defined by the C2 atoms of the lipid acyl chains.
Related collections
Most cited references72
- Record: found
- Abstract: found
- Article: not found
CHARMM-GUI: a web-based graphical user interface for CHARMM.
- Record: found
- Abstract: found
- Article: not found
Antimicrobial peptides of multicellular organisms.
- Record: found
- Abstract: found
- Article: not found
