- Record: found
- Abstract: found
- Article: found
Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review
Read this article at
Abstract
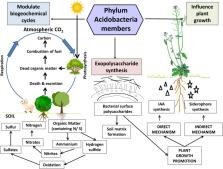
Acidobacteria represents an underrepresented soil bacterial phylum whose members are pervasive and copiously distributed across nearly all ecosystems. Acidobacterial sequences are abundant in soils and represent a significant fraction of soil microbial community. Being recalcitrant and difficult-to-cultivate under laboratory conditions, holistic, polyphasic approaches are required to study these refractive bacteria extensively. Acidobacteria possesses an inventory of genes involved in diverse metabolic pathways, as evidenced by their pan-genomic profiles. Because of their preponderance and ubiquity in the soil, speculations have been made regarding their dynamic roles in vital ecological processes viz., regulation of biogeochemical cycles, decomposition of biopolymers, exopolysaccharide secretion, and plant growth promotion. These bacteria are expected to have genes that might help in survival and competitive colonization in the rhizosphere, leading to the establishment of beneficial relationships with plants. Exploration of these genetic attributes and more in-depth insights into the belowground mechanics and dynamics would lead to a better understanding of the functions and ecological significance of this enigmatic phylum in the soil-plant environment. This review is an effort to provide a recent update into the diversity of genes in Acidobacteria useful for characterization, understanding ecological roles, and future biotechnological perspectives.
Related collections
Most cited references136

- Record: found
- Abstract: found
- Article: found
dbCAN2: a meta server for automated carbohydrate-active enzyme annotation
- Record: found
- Abstract: found
- Article: not found
Community structure and metabolism through reconstruction of microbial genomes from the environment.
- Record: found
- Abstract: found
- Article: not found