- Record: found
- Abstract: found
- Article: found
Meckel’s Cartilage in Mandibular Development and Dysmorphogenesis
Read this article at
Abstract
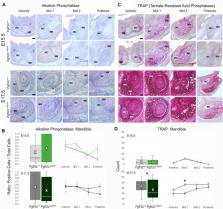
The Fgfr2c C342Y/+ Crouzon syndrome mouse model carries a cysteine to tyrosine substitution at amino acid position 342 (Cys342Tyr; C342Y) in the fibroblast growth factor receptor 2 ( Fgfr2) gene equivalent to a FGFR2 mutation commonly associated with Crouzon and Pfeiffer syndromes in humans. The Fgfr2c C342Y mutation results in constitutive activation of the receptor and is associated with upregulation of osteogenic differentiation. Fgfr2c C342Y/+ Crouzon syndrome mice show premature closure of the coronal suture and other craniofacial anomalies including malocclusion of teeth, most likely due to abnormal craniofacial form. Malformation of the mandible can precipitate a plethora of complications including disrupting development of the upper jaw and palate, impediment of the airway, and alteration of occlusion necessary for proper mastication. The current paradigm of mandibular development assumes that Meckel’s cartilage (MC) serves as a support or model for mandibular bone formation and as a template for the later forming mandible. If valid, this implies a functional relationship between MC and the forming mandible, so mandibular dysmorphogenesis might be discerned in MC affecting the relationship between MC and mandibular bone. Here we investigate the relationship of MC to mandible development from the early mineralization of the mandible (E13.5) through the initiation of MC degradation at E17.7 using Fgfr2c C342Y/+ Crouzon syndrome embryos and their unaffected littermates ( Fgfr2c +/+ ). Differences between genotypes in both MC and mandibular bone are subtle, however MC of Fgfr2c C342Y/+ embryos is generally longer relative to unaffected littermates at E15.5 with specific aspects remaining relatively large at E17.5. In contrast, mandibular bone is smaller overall in Fgfr2c C342Y/+ embryos relative to their unaffected littermates at E15.5 with the posterior aspect remaining relatively small at E17.5. At a cellular level, differences are identified between genotypes early (E13.5) followed by reduced proliferation in MC (E15.5) and in the forming mandible (E17.5) in Fgfr2c C342Y/+ embryos. Activation of the ERK pathways is reduced in the perichondrium of MC in Fgfr2c C342Y/+ embryos and increased in bone related cells at E15.5. These data reveal that the Fgfr2c C342Y mutation differentially affects cells by type, location, and developmental age indicating a complex set of changes in the cells that make up the lower jaw.
Related collections
Most cited references69
- Record: found
- Abstract: not found
- Book Chapter: not found
U-Net: Convolutional Networks for Biomedical Image Segmentation
- Record: found
- Abstract: found
- Article: not found
The Fibroblast Growth Factor signaling pathway
- Record: found
- Abstract: found
- Article: not found