- Record: found
- Abstract: found
- Article: found
Kidney injury molecule-1 inhibits metastasis of renal cell carcinoma
Read this article at
Abstract
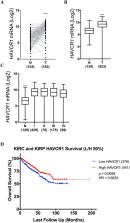
Metastasis is present in approximately 30% of patients diagnosed with renal cell carcinoma (RCC) and is associated with a 5-year survival rate of < 15%. Kidney injury molecule 1 (KIM-1), encoded by the HAVCR1 gene, is a proximal tubule cell-surface glycoprotein and a biomarker for early detection of RCC, but its pathophysiological significance in RCC remains unclear. We generated human and murine RCC cell lines either expressing or lacking KIM-1, respectively, and compared their growth and metastatic properties using validated methods. Surprisingly, KIM-1 expression had no effect on cell proliferation or subcutaneous tumour growth in immune deficient (Rag1 −/−) Balb/c mice, but inhibited cell invasion and formation of lung metastasis in the same model. Further, we show that the inhibitory effect of KIM-1 on metastases was observed in both immune deficient and immune competent mice. Transcriptomic profiling identified the mRNA for the pro-metastatic GTPase, Rab27b, to be downregulated significantly in KIM-1 expressing human and murine RCC cells. Finally, analysis of The Cancer Genome Atlas (TCGA) data revealed that elevated HAVCR1 mRNA expression in the two most common types of RCC, clear cell and papillary RCC, tumours correlated with significantly improved overall patient survival. Our findings reveal a novel role for KIM-1 in inhibiting metastasis of RCC and suggests that tumour-associated KIM-1 expression may be a favourable prognostic factor.
Related collections
Most cited references34

- Record: found
- Abstract: found
- Article: found
RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome
- Record: found
- Abstract: found
- Article: not found
Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma
- Record: found
- Abstract: found
- Article: not found