- Record: found
- Abstract: found
- Article: found
Identification of new transmembrane proteins concentrated at the nuclear envelope using organellar proteomics of mesenchymal cells
Read this article at
ABSTRACT
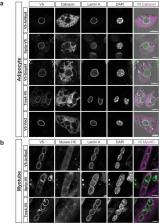
The double membrane nuclear envelope (NE), which is contiguous with the ER, contains nuclear pore complexes (NPCs) – the channels for nucleocytoplasmic transport, and the nuclear lamina (NL) – a scaffold for NE and chromatin organization. Since numerous human diseases linked to NE proteins occur in mesenchyme-derived cells, we used proteomics to characterize NE and other subcellular fractions isolated from mesenchymal stem cells and from adipocytes and myocytes. Based on spectral abundance, we calculated enrichment scores for proteins in the NE fractions. We demonstrated by quantitative immunofluorescence microscopy that five little-characterized proteins with high enrichment scores are substantially concentrated at the NE, with Itprip exposed at the outer nuclear membrane, Smpd4 enriched at the NPC, and Mfsd10, Tmx4, and Arl6ip6 likely residing in the inner nuclear membrane. These proteins provide new focal points for studying the functions of the NE. Moreover, our datasets provide a resource for evaluating additional potential NE proteins.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Sphingolipids and their metabolism in physiology and disease
- Record: found
- Abstract: found
- Article: not found
Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome.
- Record: found
- Abstract: found
- Article: not found