- Record: found
- Abstract: found
- Article: found
Structural Studies of the Lipopolysaccharide Isolated from Plesiomonas shigelloides O22:H3 (CNCTC 90/89)
Read this article at
Abstract
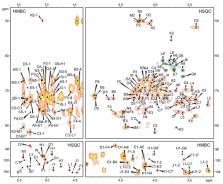
Plesiomonas shigelloides is a Gram-negative, rod-shaped bacterium which causes foodborne intestinal infections, including gastroenteritis. It is one of the most frequent causes of travellers’ diarrhoea. Lipopolysaccharide (LPS, endotoxin), an important virulence factor of the species, is in most cases characterised by a smooth character, demonstrated by the presence of all regions, such as lipid A, core oligosaccharide, and O-specific polysaccharide, where the latter part determines O-serotype. P. shigelloides LPS is still a poorly characterised virulence factor considering a “translation” of the particular O-serotype into chemical structure. To date, LPS structure has only been elucidated for 15 strains out of 102 O-serotypes. Structures of the new O-specific polysaccharide and core oligosaccharide of P. shigelloides from the Czechoslovak National Collection of Type Cultures CNCTC 90/89 LPS (O22), investigated by chemical analysis, mass spectrometry, and 1H, 13C nuclear magnetic resonance (NMR) spectroscopy, have now been reported. The pentasaccharide repeating unit of the O-specific polysaccharide is built of one d-Qui pNAc and is rich in four d-Gal pNAcAN residues. Moreover, the new core oligosaccharide shares common features of other P. shigelloides endotoxins, i.e., the lack of phosphate groups and the presence of uronic acids.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: found
NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy
- Record: found
- Abstract: not found
- Article: not found
A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates
- Record: found
- Abstract: not found
- Article: not found