- Record: found
- Abstract: found
- Article: found
Differential Expression and Roles of Staphylococcus aureus Virulence Determinants during Colonization and Disease
Read this article at
ABSTRACT
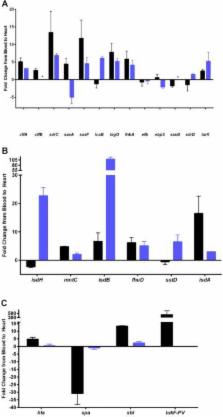
Staphylococcus aureus is a Gram-positive, commensal bacterium known to asymptomatically colonize the human skin, nares, and gastrointestinal tract. Colonized individuals are at increased risk for developing S. aureus infections, which range from mild skin and soft tissue infections to more severe diseases, such as endocarditis, bacteremia, sepsis, and osteomyelitis. Different virulence factors are required for S. aureus to infect different body sites. In this study, virulence gene expression was analyzed in two S. aureus isolates during nasal colonization, bacteremia and in the heart during sepsis. These models were chosen to represent the stepwise progression of S. aureus from an asymptomatic colonizer to an invasive pathogen. Expression of 23 putative S. aureus virulence determinants, representing protein and carbohydrate adhesins, secreted toxins, and proteins involved in metal cation acquisition and immune evasion were analyzed. Consistent upregulation of sdrC, fnbA, fhuD, sstD, and hla was observed in the shift between colonization and invasive pathogen, suggesting a prominent role for these genes in staphylococcal pathogenesis. Finally, gene expression data were correlated to the roles of the genes in pathogenesis by using knockout mutants in the animal models. These results provide insights into how S. aureus modifies virulence gene expression between commensal and invasive pathogens.
IMPORTANCE
Many bacteria, such as Staphylococcus aureus, asymptomatically colonize human skin and nasal passages but can also cause invasive diseases, such as bacteremia, pneumonia, sepsis, and osteomyelitis. The goal of this study was to analyze differences in the expression of selected S. aureus genes during a commensal lifestyle and as an invasive pathogen to gain insight into the commensal-to-pathogen transition and how a bacterial pathogen adapts to different environments within a host (e.g., from nasal colonization to invasive pathogen). The gene expression data were also used to select genes for which to construct knockout mutants to assess the role of several proteins in nasal colonization and lethal bacteremia. These results not only provide insight into the factors involved in S. aureus disease pathogenesis but also provide potential therapeutic targets.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Allelic replacement in Staphylococcus aureus with inducible counter-selection.

- Record: found
- Abstract: found
- Article: found