- Record: found
- Abstract: found
- Article: found
RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
Read this article at
Abstract
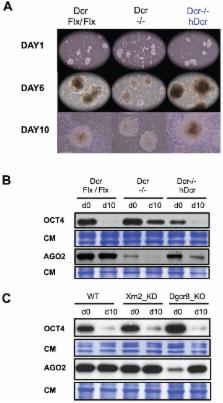
In most mouse tissues, long-interspersed elements-1 (L1s) are silenced via methylation of their 5′-untranslated regions (5′-UTR). A gradual loss-of-methylation in pre-implantation embryos coincides with L1 retrotransposition in blastocysts, generating potentially harmful mutations. Here, we show that Dicer- and Ago2-dependent RNAi restricts L1 accumulation and retrotransposition in undifferentiated mouse embryonic stem cells (mESCs), derived from blastocysts. RNAi correlates with production of Dicer-dependent 22-nt small RNAs mapping to overlapping sense/antisense transcripts produced from the L1 5′-UTR. However, RNA-surveillance pathways simultaneously degrade these transcripts and, consequently, confound the anti-L1 RNAi response. In Dicer −/− mESC complementation experiments involving ectopic Dicer expression, L1 silencing was rescued in cells in which microRNAs remained strongly depleted. Furthermore, these cells proliferated and differentiated normally, unlike their non-complemented counterparts. These results shed new light on L1 biology, uncover defensive, in addition to regulatory roles for RNAi, and raise questions on the differentiation defects of Dicer −/− mESCs.
Author Summary
A basal network of gene regulation orchestrates the processes ensuring maintenance of genome integrity. Eukaryotic small RNAs generated by the RNAse-III Dicer have emerged as central players in this network, by mediating gene silencing at the transcriptional or post-transcriptional level via RNA interference (RNAi). To gain insight into their potential developmental functions in mammals, we have characterized small RNA expression profiles during mouse Embryonic Stem Cell (mESCs) differentiation, a model for early mammalian development. Long interspersed elements 1 (L1) are non-long-terminal-repeat retrotransposons that dominate the mouse genomic landscape, and are expressed in germ cells or during early development and mESCs. Based on clear precedents in plants and fission yeast, we investigated a role for RNAi and other RNA-based pathways in the regulation of L1 transcription and mobilization. Our work uncovered the existence of small (s)RNAs that map to active L1 elements. Some have characteristics of cognate siRNA produced by Dicer, while others display strand biases and length heterogeneity that evoke their biogenesis through RNA surveillance pathways, in a Dicer-independent manner. Furthermore, genetic ablation of DICER or of ARGONAUTE proteins has complex and profound consequences on L1 transcription and mobilization, indicating that endogenous RNAi do indeed maintain genomic integrity against L1 proliferation.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs.
- Record: found
- Abstract: found
- Article: not found
MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation.
- Record: found
- Abstract: found
- Article: not found