- Record: found
- Abstract: found
- Article: not found
The Stem Cell Population of the Human Colon Crypt: Analysis via Methylation Patterns
Read this article at
Abstract
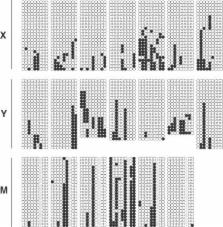
The analysis of methylation patterns is a promising approach to investigate the genealogy of cell populations in an organism. In a stem cell–niche scenario, sampled methylation patterns are the stochastic outcome of a complex interplay between niche structural features such as the number of stem cells within a niche and the niche succession time, the methylation/demethylation process, and the randomness due to sampling. As a consequence, methylation pattern studies can reveal niche characteristics but also require appropriate statistical methods. The analysis of methylation patterns sampled from colon crypts is a prototype of such a study. Previous analyses were based on forward simulation of the cell content of the whole crypt and subsequent comparisons between simulated and experimental data using a few statistics as a proxy to summarize the data. In this paper we develop a more powerful method to analyze these data based on coalescent modelling and Bayesian inference. Results support a scenario where the colon crypt is maintained by a high number of stem cells; the posterior indicates a number greater than eight and the posterior mode is between 15 and 20. The results also provide further evidence for synergistic effects in the methylation/demethylation process that could for the first time be quantitatively assessed through their long-term consequences such as the coexistence of hypermethylated and hypomethylated patterns in the same colon crypt.
Author Summary
The dynamics of the stem cell populations in human colon crypts are of interest to cancer researchers and stem cell biologists alike. One approach to studying stem cell divisions would be to adopt methods from population genetics: cells are sampled from crypts, DNA markers such as single nucleotide polymorphisms are identified, and a model of how these mutations arose is used to infer aspects of the ancestry of the sample. Because cells within an individual are being studied, mutations of this sort are extremely rare, and an alternative marker has to be used. Methylation patterns provide a feasible alternative, containing information similar to that obtained from short DNA sequences. The present study shows how such data can be used to infer aspects of stem cell dynamics, including inference about the likely number of stem cells in a crypt. In addition, biological aspects of methylation and demethylation are also studied.
Related collections
Most cited references34
- Record: found
- Abstract: not found
- Article: not found
The sampling theory of selectively neutral alleles.
- Record: found
- Abstract: not found
- Article: not found
On the Genealogy of Large Populations
- Record: found
- Abstract: found
- Article: not found
