- Record: found
- Abstract: found
- Article: not found
Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated
Read this article at
Abstract
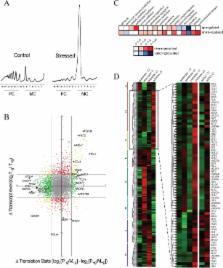
The mechanism and consequences of the translational inhibition caused by glucose depletion in yeast are characterized. eIF4A is lost from the preinitiation complex, and the pentose phosphate pathway is translationally up-regulated, allowing an efficient transition to the new conditions.
Abstract
Cellular stress can globally inhibit translation initiation, and glucose removal from yeast causes one of the most dramatic effects in terms of rapidity and scale. Here we show that the same rapid inhibition occurs during yeast growth as glucose levels diminish. We characterize this novel regulation showing that it involves alterations within the 48S preinitiation complex. In particular, the interaction between eIF4A and eIF4G is destabilized, leading to a temporary stabilization of the eIF3–eIF4G interaction on the 48S complex. Under such conditions, specific mRNAs that are important for the adaptation to the new conditions must continue to be translated. We have determined which mRNAs remain translated early after glucose starvation. These experiments enable us to provide a physiological context for this translational regulation by ascribing defined functions that are translationally maintained or up-regulated. Overrepresented in this class of mRNA are those involved in carbohydrate metabolism, including several mRNAs from the pentose phosphate pathway. Our data support a hypothesis that a concerted preemptive activation of the pentose phosphate pathway, which targets both mRNA transcription and translation, is important for the transition from fermentative to respiratory growth in yeast.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
Exploring the metabolic and genetic control of gene expression on a genomic scale.
- Record: found
- Abstract: found
- Article: not found
Regulation of cap-dependent translation by eIF4E inhibitory proteins.
- Record: found
- Abstract: found
- Article: not found
